How to Read Mat Files in R and Access Struct Fields
How to load Matlab .mat files in Python
![]()

Matlab is a really popular platform for scientific computing in the academia. I've used information technology my throughout my engineering degree and chances are, you will come beyond .mat files for datasets released by the universities.
This is a brief post which explains how to load these files using python, the virtually popular language for car learning today.
The information
I wanted to build a classifier for detectin 1000 cars of unlike models and makes and so the Stanford Cars Dataset appeared to be a great starting bespeak. Coming from the academia, the annotations for the dataset was in the .mat format. You can get the file used in this mail service here.
Loading .mat files
Scipy is a really popular python library used for scientific calculating and quite naturally, they take a method which lets you read in .mat files. Reading them in is definitely the piece of cake part. Yous tin can get it done in one line of lawmaking:
from scipy.io import loadmat
annots = loadmat('cars_train_annos.mat') Well, it's actually that simple. Simply permit's go on and actually try to go the data we need out of this lexicon.
Formatting the data
The loadmat method returns a more than familiar data structure, a python dictionary. If we peek into the keys, we'll run into how at home we feel now compared to dealing with a .mat file:
annots.keys()
> dict_keys(['__header__', '__version__', '__globals__', 'annotations']) Looking at the documentation for this dataset, we'll get to learn what this is really fabricated of. The README.txt gives u.s.a. the post-obit information:
This file gives documentation for the cars 196 dataset.
(http://ai.stanford.edu/~jkrause/cars/car_dataset.html) — — — — — — — — — — — — — — — — — — — —
Metadata/Annotations
— — — — — — — — — — — — — — — — — — — —
Descriptions of the files are as follows: -cars_meta.mat:
Contains a cell array of class names, one for each grade. -cars_train_annos.mat:
Contains the variable 'annotations', which is a struct assortment of length
num_images and where each element has the fields:
bbox_x1: Min 10-value of the bounding box, in pixels
bbox_x2: Max x-value of the bounding box, in pixels
bbox_y1: Min y-value of the bounding box, in pixels
bbox_y2: Max y-value of the bounding box, in pixels
course: Integral id of the class the image belongs to.
fname: Filename of the image inside the binder of images. -cars_test_annos.mat:
Aforementioned format as 'cars_train_annos.mat', except the class is not provided. — — — — — — — — — — — — — — — — — — — —
Submission file format
— — — — — — — — — — — — — — — — — — — —
Files for submission should be .txt files with the class prediction for
paradigm Thou on line Grand. Note that epitome M corresponds to the Mth notation in
the provided notation file. An example of a file in this format is
train_perfect_preds.txt Included in the devkit are a script for evaluating training accuracy,
eval_train.thou. Usage is: (in MATLAB)
>> [accuracy, confusion_matrix] = eval_train('train_perfect_preds.txt') If your training predictions work with this function then your testing
predictions should exist practiced to go for the evaluation server, assuming
that they're in the same format equally your training predictions.
Our involvement is in the 'annotations' variable, equally it contains our class labels and bounding boxes. Information technology's a struct, a information type very familiar to folks coming from a strongly typed language like a season of C or coffee.
A little excavation into the object gives us some interesting things to work with:
type(annots['annotations']),annots['annotations'].shape
>(numpy.ndarray, (ane, 8144)) blazon(annots['annotations'][0][0]),annots['annotations'][0][0].shape
>(numpy.void, ())
The annotations are stored in a numpy.ndarray format, however the data type for the items inside this array is numpy.void and numpy doesn't really seem to know the shape of them.
The documentation page for the loadmat method tells us how it loads matlab structs into numpy structured arrays.You tin access the members of the structs using the keys:
annots['annotations'][0][0]['bbox_x1'], annots['annotations'][0][0]['fname'] > (array([[39]], dtype=uint8), assortment(['00001.jpg'], dtype='<U9'))
Then now that we know how to access the members of the struct, we tin can iterate through all of them and store them in a list:
[particular.flat[0] for particular in annots['annotations'][0][0]] > [39, 116, 569, 375, xiv, '00001.jpg']
Here, nosotros tin can use the flat method to squeeze the value out of the array.
Hullo Pandas
Now that we know how to deal with matlab files in python, allow's convert it into a pandas information frame. Nosotros tin can do then hands using a list of lists:
information = [[row.flat[0] for row in line] for line in annots['annotations'][0]] columns = ['bbox_x1', 'bbox_y1', 'bbox_x2', 'bbox_y2', 'class', 'fname']
df_train = pd.DataFrame(data, columns=columns)
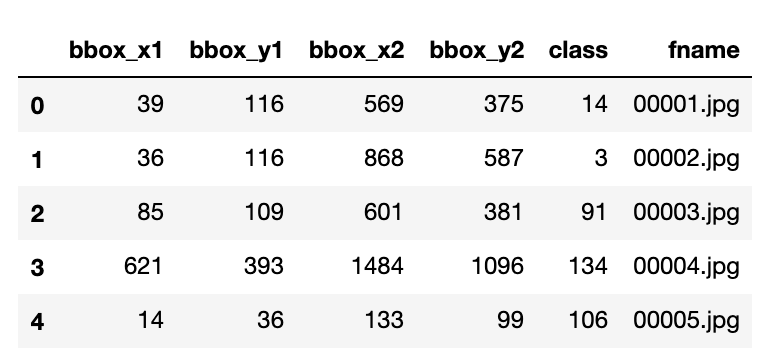
Finally, familiar territory!
The code for this mail service can exist establish hither.
Source: https://towardsdatascience.com/how-to-load-matlab-mat-files-in-python-1f200e1287b5
0 Response to "How to Read Mat Files in R and Access Struct Fields"
Post a Comment